Showing 11 to 20 of 29 entries
Number of Genomic Records: 242607
Statewise SARS-CoV-2 Sequences
Gender Distribution
Age Group Distribution
SARS-CoV-2 Different Clade: Monthly Distribution
Monthly GISAID Clade |
Monthly Nextstrain Clade |
Monthly Pangolin Clade |
Monthly Group Clade |
SARS-CoV-2 Genome Variants References Table
| Sr No. | Lineage Group | Lineage Name |
|---|---|---|
| 11 | BA.4 | BA.4, BA.4.1, BA.4.1.1, BA.4.2, BA.4.4, BA.4.6, BA.4.1.8, BA.4.7, BA.4.1.5, BA.4.8, BA.4.1.9, BA.4.6.1, BA.4.1.6, BA.4.6.3, BA.4.6.5 |
| 12 | BA.5 | BA.5, BA.5.2.1, BA.5.2, BA.5.1, BA.5.3.1, BA.5.5, BE.1, BF.1, BA.5.1.3, BE.1.1, BE.3, BF.2, BF.3, BF.4, BA.5.1.4, BA.5.1.2, BA.5.3, BA.5.6, BA.5.2.3, BA.5.1.1, BF.5, BA.5.3.3, BA.5.2.4, BF.10, BF.3.1, BF.8, BA.5.2.2, BA.5.1.10, BF.12, BA.5.8, BA.5.2.26, BA.5.2.6, BA.5.2.21, BA.5.1.24, BF.23, BA.5.1.12, BA.5.1.25, BF.27, BA.5.2.27, BF.21, BF.24, BA.5.2.28, BA.5.1.17, BA.5.2.8, BF.20, BE.4.1, BA.5.1.23, BF.26, BA.5.1.22, BA.5.2.31, BF.15, CE.1, BA.5.2.30, BA.5.2.22, BA.5.2.33, BA.5.2.18, BF.28, BA.5.2.19, BA.5.2.16, BA.5.2.24, BA.5.2.9, BE.1.1.2, BF.18, BA.5.2.20, BE.4, BF.6, BA.5.9, BF.11.5, CL.1, BE.1.2, BA.5.1.18, CK.1, BA.5.2.34, CK.3, BA.5.1.28, BF.34, BF.31, CR.1, BA.5.2.42, BF.14, BA.5.2.25, BA.5.1.8, BA.5.2.12, CP.1, BE.1.4, BM.3, BV.2, CP.6, BA.5.2.38, BA.5.2.36, BA.5.1.30, BE.5, BA.5.11, CR.2, BA.5.2.44, BA.5.2.37, BA.5.2.41, BA.5.1.15, BA.5.2.48, BW.1.1, DF.1, EF.1, BA.5.1.29, BF.38, BA.5.1.35, EC.1.1, BF.7.22, EF.1.1.1, DU.1, BA.5.2.56, BF.7.14, BF.7.8, BA.5.2.47, FC.1, EE.2, BA.5.2.59, DY.4, BF.41, BA.5.2.49, BA.5.1.36, BE.12, DY.2, BA.5.2.62, BF.36, CP.7, FN.1, DN.3, DN.2, BE.1.1.1, CK.1.5, BU.1 |
| 13 | Beta | B.1.351, B.1.351.2, B.1.351.3 |
| 14 | BF.7 | BF.7, BF.7.12, BF.7.5.1, BF.7.4.1, BF.7.4, BF.7.6, BF.7.15, BF.7.14.6 |
| 15 | BQ | BQ.1, BQ.1.5, BQ.1.14, BQ.1.1, BQ.1.1.5, BQ.1.9, BQ.1.1.13, BQ.1.1.15, BQ.1.2, BQ.1.1.4, BQ.1.1.18, BQ.1.23, BQ.1.1.7, BQ.1.25, BQ.1.1.1, BQ.1.12, BQ.1.10, BQ.1.8, BQ.1.1.22, BQ.1.19, BQ.1.1.3, BQ.1.8.2, BQ.1.13, BQ.1.15, BQ.1.10.1, BQ.1.24, BQ.1.1.32, BQ.1.1.2, BQ.1.1.45, BQ.1.3, BQ.1.1.52, BQ.1.1.35, BQ.1.13.1, BQ.1.1.8, BQ.1.1.38, BQ.1.1.64, BQ.1.1.47, BQ.1.1.59, BQ.1.2.1, BQ.1.1.41, BQ.1.1.11, BQ.1.1.48, BQ.1.1.68, BQ.1.1.69, BQ.1.1.51, BQ.1.1.24, BQ.1.1.31, BQ.1.2.3, BQ.1.1.67, BQ.1.1.74, BQ.1.3.2 |
| 16 | Delta | AY.1, AY.113, AY.122, B.1.617.2, AY.100, AY.101, AY.102, AY.103, AY.104, AY.105, AY.106, AY.107, AY.108, AY.109, AY.110, AY.111, AY.112, AY.112.2, AY.116, AY.117, AY.118, AY.119, AY.119.2, AY.120, AY.120.2, AY.121.1, AY.122.1, AY.122.6, AY.124, AY.124.1.1, AY.125, AY.126, AY.127, AY.127.1, AY.128, AY.129, AY.131, AY.15, AY.16, AY.20, AY.21, AY.23, AY.23.1, AY.24, AY.25, AY.25.1, AY.27, AY.28, AY.29.1, AY.3, AY.30, AY.32, AY.33, AY.34, AY.34.1, AY.36, AY.37, AY.38, AY.39, AY.39.1, AY.39.1.1, AY.39.1.4, AY.39.3, AY.4, AY.4.13, AY.42, AY.4.2, AY.4.2.1, AY.4.2.2, AY.4.2.3, AY.43, AY.4.3, AY.43.7, AY.44, AY.45, AY.4.5, AY.46, AY.46.2, AY.46.3, AY.46.4, AY.46.5, AY.46.6, AY.47, AY.48, AY.4.8, AY.5, AY.52, AY.53, AY.54, AY.5.4, AY.55, AY.57, AY.58, AY.59, AY.6, AY.61, AY.65, AY.71, AY.75, AY.76, AY.77, AY.79, AY.8, AY.80, AY.82, AY.83, AY.84, AY.85, AY.86, AY.88, AY.9, AY.91, AY.92, AY.93, AY.94, AY.95, AY.98, AY.98.1, AY.99, AY.99.1, AY.120.1, AY.121, AY.29, AY.39.1.2, AY.4.7, AY.4.9, AY.5.3, AY.33.2, AY.35, AY.70, AY.4.11 |
| 17 | Epsilon | B.1.429 |
| 18 | Eta | B.1.525 |
| 19 | Gamma | P.1, P.1.1 |
| 20 | Iota | B.1.526 |
SARS-CoV-2 Genome Variants Study References
Nextstrain Clade
Source: Nextstrain Clade (https://nextstrain.org/)
SARS-CoV-2 Genome Protein
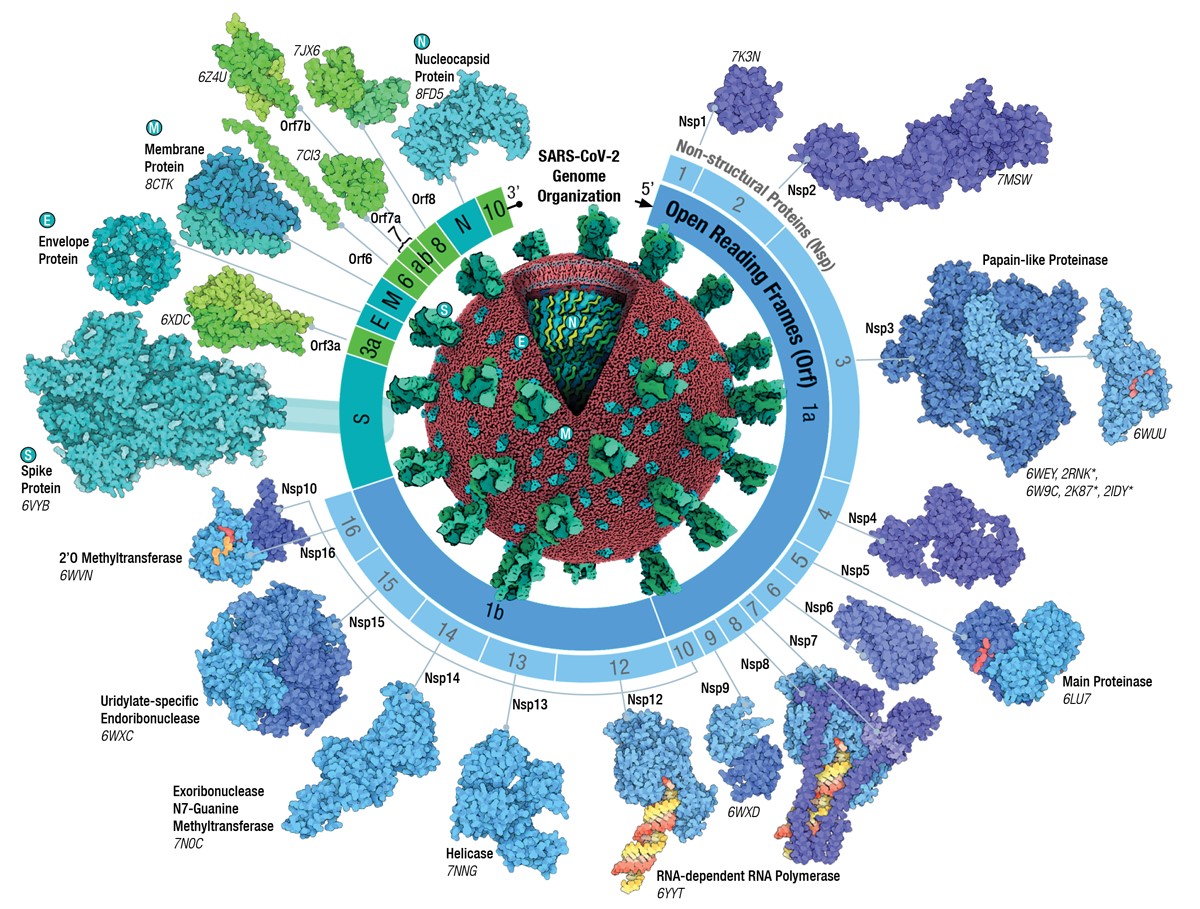
Source: SARS-CoV-2 Genome Protein (RCSB.org/covid19)
Our Team
| Name | Position | |
|---|---|---|
| Dr Pratip Shil | Principal investigator (PI) | |
| Dr Sarah S Cherian | Co-investigator (Co-I) | |
| Dr Varsha A Potdar | Co-investigator (Co-I) | |
| Mr Santoshkumar Jadhav | Sr Statistician | Statistical Data Analysis |
| Dr Nitin M Atre | Project Research Scientist-V | Dashboard Design, Development and Programmer |
| Mr Ashokkumar J | Project Technical Support-II | Bioinformatics Data Analysis |
Acknowledgment
This project is funded by Indian Council of Medical Research (ICMR), New Delhi (Project ID: 2021-6366)
We are really thankful to all our data sources
We are really thankful to all our data sources
- Indian SARS-CoV-2 Genomics Consortium (INSACOG)
- GISAID
- Nextstrain
- Pangolin