Showing 21 to 29 of 29 entries
Number of Genomic Records: 242607
Statewise SARS-CoV-2 Sequences
Gender Distribution
Age Group Distribution
SARS-CoV-2 Different Clade: Monthly Distribution
Monthly GISAID Clade |
Monthly Nextstrain Clade |
Monthly Pangolin Clade |
Monthly Group Clade |
SARS-CoV-2 Genome Variants References Table
| Sr No. | Lineage Group | Lineage Name |
|---|---|---|
| 21 | JN.1 | JN.1, JN.1.4, JN.1.1.3, JN.1.10, JN.1.3, JN.1.6, JN.1.1.1, JN.1.1, JN.1.8, JN.1.2, JN.1.5, JN.1.9, JN.1.11, JN.1.19, JN.1.11.1, JN.1.18, JN.1.4.2, JN.1.20, JN.1.9.1, JN.1.15, JN.1.22, JN.1.16, JN.1.1.5, JN.1.25.1, JN.1.25, JN.1.13, JN.1.7.2, JN.1.1.6, JN.1.1.7, JN.1.16.1, JN.1.18.1, JN.1.28, JN.1.28.1, JN.1.29, JN.1.30, JN.1.30.1, JN.1.32, JN.1.33, JN.1.37, JN.1.38, JN.1.39, JN.1.4.4, JN.1.4.5, JN.1.4.7, JN.1.40, JN.1.43.1, JN.1.44, JN.1.47, JN.1.48, JN.1.48.1, JN.1.8.3, JN.1.9.2, KU.1, KU.2, KZ.1, KZ.1.1, LB.1, LC.1, JN.1.6.1, JN.1.1.10, JN.1.1.8, JN.1.18.3, JN.1.34, JN.1.39.2, JN.1.39.3, JN.1.42, JN.1.44.1, JN.1.46, JN.1.48.2, JN.1.49, JN.1.49.1, JN.1.49.2, JN.1.52, JN.1.53, JN.1.57, JN.1.58, JN.1.60, JN.1.61, JN.1.63.1, KQ.1, LB.1.2, LB.1.2.1, LB.1.3, LB.1.4, LM.1, LQ.1 |
| 22 | Kappa | B.1.617.1 |
| 23 | KP.x | KP.2, KP.1.1, KP.1, KP.1.1.1, KP.1.2, KP.2.1, KP.2.2, KP.2.3, KP.3, KP.4, KP.4.1, KP.4.2, KP.1.1.3, KP.1.1.5, KP.2.16, KP.2.4, KP.2.5, KP.2.7, KP.3.1, KP.3.1.4, KP.3.2, KP.3.2.1, KP.4.1.1, KP.4.2.1, KP.5, KP.2.15 |
| 24 | Omicron | B.1.1.529, XAB, XAH, XE, XN, XAG, XAP, XAS, XAQ, XAM, XM, XU, XBH, XBF, XAY.2, XBN, XBC.1.1, XBF.8, XBM, XBF.5, BC.1, XBL.1, XAY.2.3, XCG, XBC.1.1.2, BA.2.86.1, BA.2.86, JN.2, JN.6, EW.2, XCW, XDA, XDB, XDD, XDK, JN.12, BA.2.87, XDK.1 |
| 25 | Other | A, A.1, A.19, A.2, A.23.1, A.27, A.29, AE.3, AE.4, B, B.1, B.11, B.1.1, B.1.1.10, B.1.1.101, B.1.111, B.1.1.112, B.1.1.117, B.1.113, B.1.1.132, B.1.116, B.1.1.161, B.1.1.168, B.1.1.174, B.1.1.189, B.1.1.201, B.1.1.214, B.1.1.216, B.1.1.220, B.1.1.222, B.1.1.25, B.1.1.27, B.1.1.273, B.1.1.274, B.1.1.282, B.1.1.285, B.1.1.306, B.1.1.312, B.1.1.317, B.1.1.318, B.1.1.326, B.1.1.339, B.1.1.353, B.1.1.354, B.1.1.357, B.1.1.359, B.1.1.368, B.1.137, B.1.1.397, B.1.1.416, B.1.1.420, B.1.143, B.1.1.441, B.1.145, B.1.1.45, B.1.1.46, B.1.1.487, B.1.1.523, B.1.1.525, B.1.1.526, B.1.153, B.1.1.53, B.1.1.54, B.1.1.57, B.1.1.63, B.1.170, B.1.177, B.1.177.16, B.1.177.19, B.1.177.4, B.1.177.7, B.1.177.87, B.1.179, B.1.1.8, B.1.182, B.1.184, B.1.192, B.1.195, B.1.2, B.1.201, B.1.206, B.1.210, B.1.214.3, B.1.222, B.1.234, B.1.238, B.1.239, B.1.241, B.1.243, B.1.247, B.1.250, B.1.258, B.1.258.20, B.1.260, B.1.36, B.1.36.10, B.1.36.16, B.1.36.18, B.1.36.19, B.1.36.22, B.1.36.24, B.1.36.29, B.1.36.31, B.1.36.35, B.1.36.38, B.1.36.39, B.1.36.7, B.1.36.8, B.1.369, B.1.378, B.1.380, B.1.384, B.1.390, B.1.397, B.1.398, B.1.399, B.1.401, B.1.428, B.1.436, B.1.441, B.1.459, B.1.465, B.1.466.1, B.1.466.2, B.1.470, B.1.473, B.1.523, B.1.524, B.1.533, B.1.537, B.1.538, B.1.540, B.1.548, B.1.551, B.1.558, B.1.560, B.1.578, B.1.602, B.1.609, B.1.617, B.1.617.3, B.1.618, B.1.620, B.1.633, B.1.640.2, B.1.78, B.1.81, B.1.91, B.23, B.26, B.3, B.4, B.40, B.4.7, B.6, B.6.1, B.6.6, C.17, C.36, C.36.3, C.38, L.3, R.1, B.1.1.172, B.1.118, B.1.1.462, B.1.1.1, B.1.1.261, B.1.159, B.1.36.17, B.1.1.83, B.1.199, XC, B.55, B.1.22, B.1.240, B.1.1.164, B.1.1.28, B.1.1.166, B.1.1.89, B.1.1.355, AV.1, B.1.564, B.1.124, B.1.1.142, B.1.395, B.1.215, B.1.1.122, B.1.104, B.1.1.59, B.1.1.294, B.1.1.38, B.1.466, B.1.1.135, B.1.1.254, B.1.214, B.1.468, B.1.518, B.1.237, B.1.1.33 |
| 26 | XBB | XBB, XBB.1, XBB.2, XBB.3, XBB.4, XBB.5, XBB.3.1, XBB.1.3, XBB.1.4, XBB.1.5, XBB.1.1, XBB.1.9, XBB.1.2, XBB.1.9.1, XBB.2.1, XBL, XBB.1.9.2, XBB.1.11.1, XBB.2.4, XBB.1.5.5, XBB.1.5.8, XBB.1.5.7, XBB.1.5.4, XBB.1.11, XBP, XBB.1.13, XBB.1.5.1, EG.1, XBB.1.5.15, XBB.1.5.39, XBB.1.5.18, XBB.1.5.12, XBB.1.5.3, XBB.1.5.32, XBB.1.22.1, XBB.1.5.13, XBB.1.22, XBB.1.5.23, XBB.1.5.28, XBB.1.5.33, XBB.1.5.25, XBB.1.22.2, XBB.1.5.37, FD.2, XBB.1.5.38, XBB.1.5.16, XBB.1.5.14, XBB.1.5.24, XBB.6, XBB.2.8, XBB.8, XBB.1.5.11, XBB.2.7.1, EL.1, XBB.1.8, XBB.1.12, XBB.1.24, XBB.1.17.1, XBB.1.5.17, XBB.2.7, XBB.1.19.1, XBB.1.5.31, XBB.2.5, XBB.1.19, XBB.3.3, XBB.1.5.20, XBB.2.6, XBB.1.28, XBB.1.15, XBB.1.27, XBB.1.5.21, XBB.1.9.4, XBB.1.9.5, XBB.1.7, XBB.1.5.36, XBB.1.5.26, XBB.3.2, XBB.1.9.3, FD.1, EU.1.1, XBB.1.5.45, EG.2, XBB.1.5.67, EG.5, GE.1, XBB.1.30, FL.3.3, GA.3, FT.1, XBB.1.5.57, FY.1, XBB.1.34, XBB.9, FL.12, FL.4, XBB.1.5.47, XBB.1.39, EG.4, FL.1, XBB.1.5.54, XBB.1.36, FL.5, XBB.1.5.42, XBB.1.5.43, XBB.1.24.1, XBB.1.5.46, FL.10, XBB.1.5.48, FL.2.1, FL.8, FL.2, XBB.1.5.52, XBB.1.35, EU.1.1.1, XBB.3.4, XBB.1.5.56, FL.1.3, XBB.1.37.1, XBB.1.5.65, FD.4, FY.2, XBB.1.5.51, EG.5.1, EK.3, XBB.1.5.30, EG.1.2, FE.1.1, XBB.1.5.87, GJ.1.1, XBB.1.31, FY.4.1, GS.1, XBB.2.10, XBB.1.5.75, GR.1, XBB.1.34.2, XBB.1.5.66, FP.2, FP.3, XBB.1.45.1, XBB.2.12, FY.5, XBB.2.11, XBB.2.11.1, XBB.1.5.76, GF.1, EG.6, XBB.2.6.2, EG.5.2, FL.1.5.1, GJ.1, XBB.1.43.1, XBB.1.5.80, FL.13, XBB.1.42, XBB.1.43, FL.17, FP.4, FY.1.1, XBB.1.5.2, FL.4.1, FP.2.1, XBB.1.5.72, FL.15, EG.5.1.1, XBB.1.44, GK.1, XBB.1.5.91, GD.2, EG.1.6, FD.4.1, FL.4.7, XBB.1.5.59, FL.22, GJ.1.2, FL.23, FL.26, XBB.1.5.96, XBB.1.5.99, GK.1.3, GD.3, GZ.1, EG.12, FL.13.2, GW.4, XBB.1.31.2, HH.2, GA.6, GJ.4, XBB.1.19.2, HH.1, GJ.3, FL.13.3, XBB.1.49, HK.2, FY.3.1, FL.31, EG.13, GE.1.1, FL.4.11, GS.2, EG.14, FL.2.6, XCH.1, GM.3, JG.3, GW.5.1.1, FL.4.8, GW.5, HK.3, GE.1.3, EG.5.1.6, HV.1, GW.5.1, XBB.1.41.1, GK.1.1, EG.5.1.8, HK.3.1, JD.1.1, EG.5.1.15, FL.13.4, FL.2.7, FL.39, FY.9, GA.7, GJ.6, GW.5.3.1, HH.3, HH.4, HH.6, HH.7, HK.29, HK.3.5, HN.5, HV.1.11, JK.1, KB.3, XBB.1.5.110, XBB.1.5.95, JD.1.1.1, XBB.1.28.1, XBB.1.5.73, HN.3, FY.3 |
| 27 | XBB.1.16 | XBB.1.16, XBB.1.16.1, XBB.1.16.5, XBB.1.16.2, FU.1, FU.2, XBB.1.16.3, XBB.1.16.4, XBB.1.16.6, XBB.1.16.8, XBB.1.16.11, XBB.1.16.10, XBB.1.16.7, XBB.1.16.20, GY.8, GY.6, XBB.1.16.22, FU.5, XBB.1.16.21, XBB.1.16.17, GY.2, XBB.1.16.14, FU.4, XBB.1.16.16, HF.1, FL.18, FU.3, XBB.1.16.19, XBB.1.16.13, GY.4, XBB.1.16.18, XBB.1.16.12, FU.3.1, XBB.1.16.9, XBB.1.16.24, XBB.1.16.15, JM.2, JF.1, FU.1.1, JF.1.1, JG.3.2, XBB.1.16.25, XBB.1.16.26, XBB.1.16.31, GY.3, XBB.1.16.23 |
| 28 | XBB.2.3 | XBB.2.3, XBB.2.3.2, XBB.2.3.1, XBB.2.3.5, XBB.2.3.3, XBB.2.3.4, XBB.2.3.11, XBB.2.3.7, XBB.2.3.9, XBB.2.3.6, XBB.2.3.8, XBB.2.3.10, XBB.2.3.12, XBB.2.3.14, XBB.2.3.13, JE.1, JE.1.1, JE.1.1.1, JY.1, JY.1.1, XBB.2.3.18, XBB.2.3.19, XBB.2.3.20 |
| 29 | Zeta | P.2 |
SARS-CoV-2 Genome Variants Study References
Nextstrain Clade
Source: Nextstrain Clade (https://nextstrain.org/)
SARS-CoV-2 Genome Protein
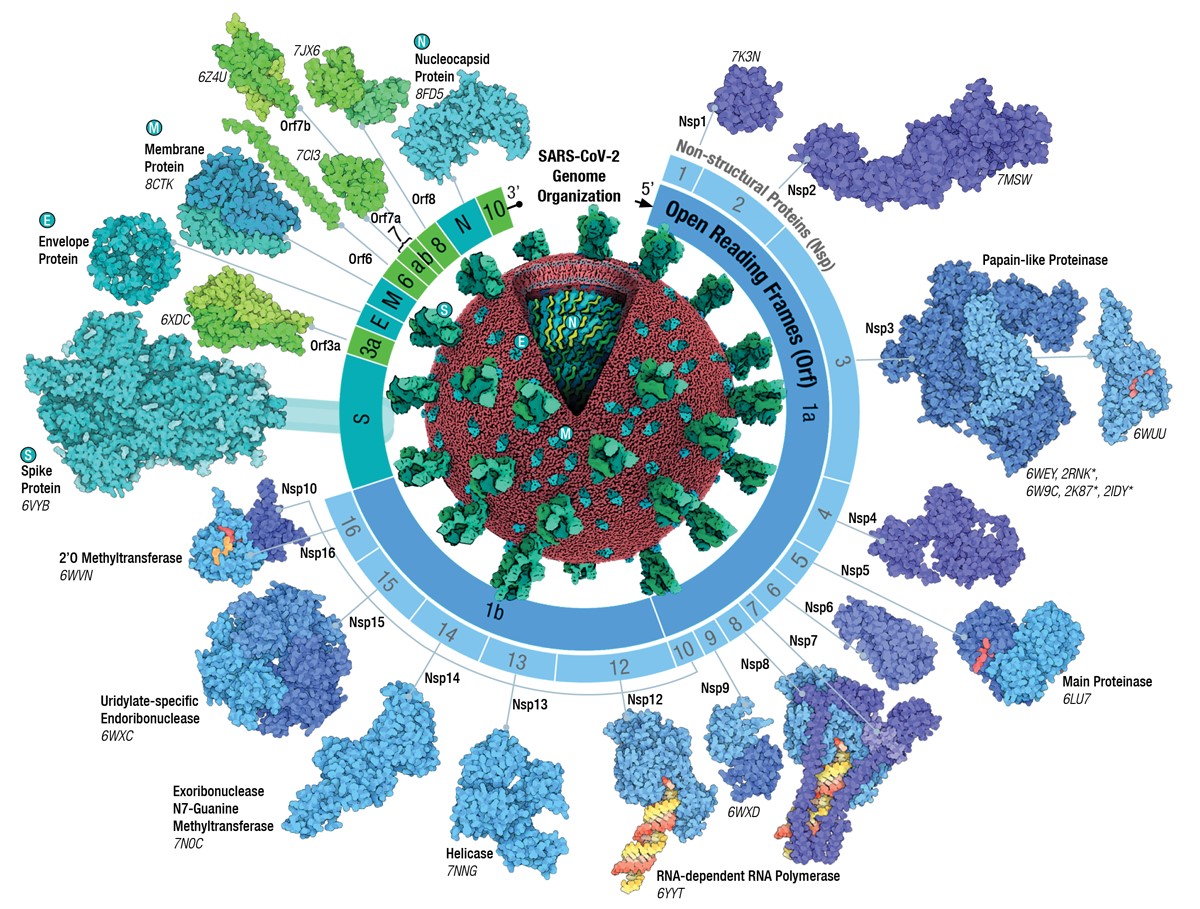
Source: SARS-CoV-2 Genome Protein (RCSB.org/covid19)
Our Team
| Name | Position | |
|---|---|---|
| Dr Pratip Shil | Principal investigator (PI) | |
| Dr Sarah S Cherian | Co-investigator (Co-I) | |
| Dr Varsha A Potdar | Co-investigator (Co-I) | |
| Mr Santoshkumar Jadhav | Sr Statistician | Statistical Data Analysis |
| Dr Nitin M Atre | Project Research Scientist-V | Dashboard Design, Development and Programmer |
| Mr Ashokkumar J | Project Technical Support-II | Bioinformatics Data Analysis |
Acknowledgment
This project is funded by Indian Council of Medical Research (ICMR), New Delhi (Project ID: 2021-6366)
We are really thankful to all our data sources
We are really thankful to all our data sources
- Indian SARS-CoV-2 Genomics Consortium (INSACOG)
- GISAID
- Nextstrain
- Pangolin